Elemental maps to dye for: using Energy Dispersive X-ray Spectrometry for quantitative and qualitative analysis of heavy metal stains.
- Abstract number
- 193
- Presentation Form
- Poster
- DOI
- 10.22443/rms.mmc2023.193
- Corresponding Email
- [email protected]
- Session
- Poster Session One
- Authors
- Mr Pedro Machado (1), Dr Errin Johnson (2), Dr Louise Hughes (1)
- Affiliations
-
1. 1. Oxford Instruments NanoAnalysis
2. 2. The William Dunn School of Pathology, The University of Oxford
- Keywords
Energy Dispersive X-ray Spectrometry, EDS, electron microscopy, EM, sample preparation, elemental maps, heavy metals, stains
- Abstract text
Biological samples are composed of elements with a low atomic number. Stains containing heavy metals are used for contrast in conventional electron microscopy by increasing the scattering of the electron beam. Many different staining methods have been developed over the years to achieve high and uniform contrast for large tissue volumes (volume electron microscopy - vEM) and to localise or label features of interest (including multimodal imaging).
For better understanding of how the contrasting elements are distributed when using different workflows, energy dispersive x-ray spectrometry (EDS) was used for an in-depth analysis and quantification of elements bound to our samples. EDS allows for the mapping and quantification of the exogenous stains, as well as the native elements in biological material. We investigated how the specimen preparation, particularly the staining workflow, is influencing the localisation of native elements as well as the elements used in the staining reagents.
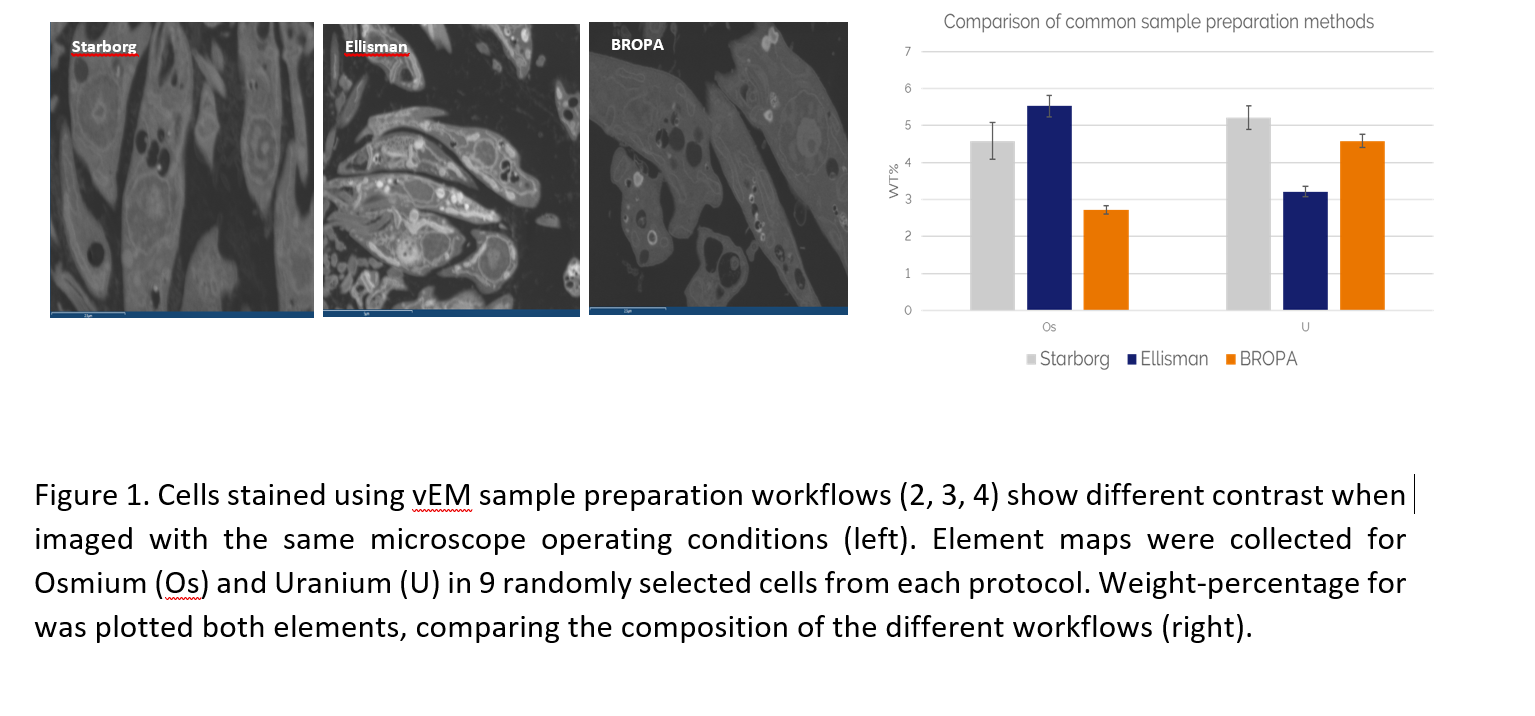
We used EDS to map and quantify the elemental variations in samples that were prepared without exogenous stains, and with a selection of different methods previously published for biological workflows that cover a wide range of applications – conventional preparation, multimodal/correlative preparations, and vEM preparations (1, 2, 3, 4, 5). The samples were sectioned at 200-300nm, collected on carbon coated Cu grids and mounted in a STEM holder for imaging. The sections were analysed using an Ultim Extreme EDS detector (typical microscope operating conditions of 5kV and a beam current of 300pA). We were able to characterise differential staining of multiple subcellular structures by collecting point spectra, elemental maps, and comparing quantitative measures of those heavy elements.
We generated a comparative analysis of the staining and native components of cells and tissues processed for electron microscopy (Fig. 1). Our data suggests that there are complex interactions between the staining agents and samples. Variations to the sample preparation workflows modulate the quantity and distribution of heavy metal stains. Surprisingly, EDS quantification does not directly correlate with the concentration of stains used during the specimen preparation. In addition, the localisation of different elements within different subcellular compartments is also dependent on the preparation workflow. EDS can provide an essential analysis tool for selecting and quality control of your specimen preparation.
Using analytical tools such as EDS, we can shed light on how the contrast of the samples can vary and impact data interpretation. This type of approach will be key for future development and standardisation of specimen preparation methods for advanced) imaging workflows.
- References
- Hua Y, Laserstein P, Helmstaedter M. Large-volume en-bloc staining for electron microscopy-based connectomics. Nat Commun. 2015 Aug 3;6:7923. doi: 10.1038/ncomms8923. PMID: 26235643; PMCID: PMC4532871.
- Starborg T, Kalson NS, Lu Y, Mironov A, Cootes TF, Holmes DF, Kadler KE. Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization. Nat Protoc. 2013;8(7):1433-48. doi: 10.1038/nprot.2013.086. Epub 2013 Jun 27. PMID: 23807286; PMCID: PMC5642902.
- Deerinck, T.J., Bushong, E.A., Thor, A.K., Ellisman, M., Deerinck, T.J., Bushong, E.A., Ellisman, M., Deerinck, T.J., & Thor, C. (2010). NCMIR methods for 3D EM: a new protocol for preparation of biological specimens for serial block face scanning electron microscopy.
- Mikula S, Denk W. High-resolution whole-brain staining for electron microscopic circuit reconstruction. Nat Methods. 2015 Jun;12(6):541-6. doi: 10.1038/nmeth.3361. Epub 2015 Apr 13. PMID: 25867849.
- Ronchi P, Mizzon G, Machado P, D'Imprima E, Best BT, Cassella L, Schnorrenberg S, Montero MG, Jechlinger M, Ephrussi A, Leptin M, Mahamid J, Schwab Y. High-precision targeting workflow for volume electron microscopy. J Cell Biol. 2021 Sep 6;220(9):e202104069. doi: 10.1083/jcb.202104069. Epub 2021 Jun 23. PMID: 34160561; PMCID: PMC8225610