A comprehensive serial block-face scanning electron microscopy solution with exchangeable acquisition system and data processing tools
- Abstract number
- 198
- Presentation Form
- Poster
- DOI
- 10.22443/rms.mmc2023.198
- Corresponding Email
- [email protected]
- Session
- Poster Session Two
- Authors
- Martin Koban (1), Markéta Machálková (2), Jakub Javůrek (2), Tomáš Borůvka (1)
- Affiliations
-
1. TESCAN Brno s.r.o.
2. TESCAN ORSAY HOLDING, a.s.
- Keywords
serial block-face SEM, volume EM, 3D data processing
- Abstract text
Techniques of volume electron microscopy (EM) have become a strong asset in scientific arsenal when three-dimensional (3D) ultrastructure of biological samples is to be examined. Serial block-face scanning electron microscopy (SBF-SEM) is one of the prominent volume EM methods and has seen a great increase in implementation across research labs, facilities and manufacturers [1]. However, there are still bottlenecks in SBF-SEM associated mainly with complex sample preparation protocols and analysis of vast amounts of generated data [2]. Due to numerous complicated steps, it is usually not possible to execute the entire SBF-SEM experiment using tools from a single vendor and the researchers are forced to laboriously assemble their own customized workflows. Another issue is exchangeability of SBF-SEM systems – some solutions require heavy reconfiguration of the SEM to switch between SBF imaging and standard operation. This makes the microscope a narrowly specialized tool, which is often incompatible with the needs of modern research facilities. Taking these pain points into consideration, we present a lightweight, easy-to-exchange SBF-SEM add-on for TESCAN SEMs, including a dedicated sensitive BSE detector and a software module for volume EM data processing, thus providing tools for smooth execution of complex SBF-SEM experiments.
TESCAN solution for SBF-SEM is based on the integration of in-chamber ultramicrotome Katana by ConnectomX Ltd. [3]. This compact device is installed on a standard SEM stage through a customized adapter which ensures superior mechanical and electrical stability – see Figure 1. The microtome and the adapter can be easily (un)mounted from the stage and replaced with the standard sample holder, an operation that can be performed by a regular microscope user in the order of minutes.
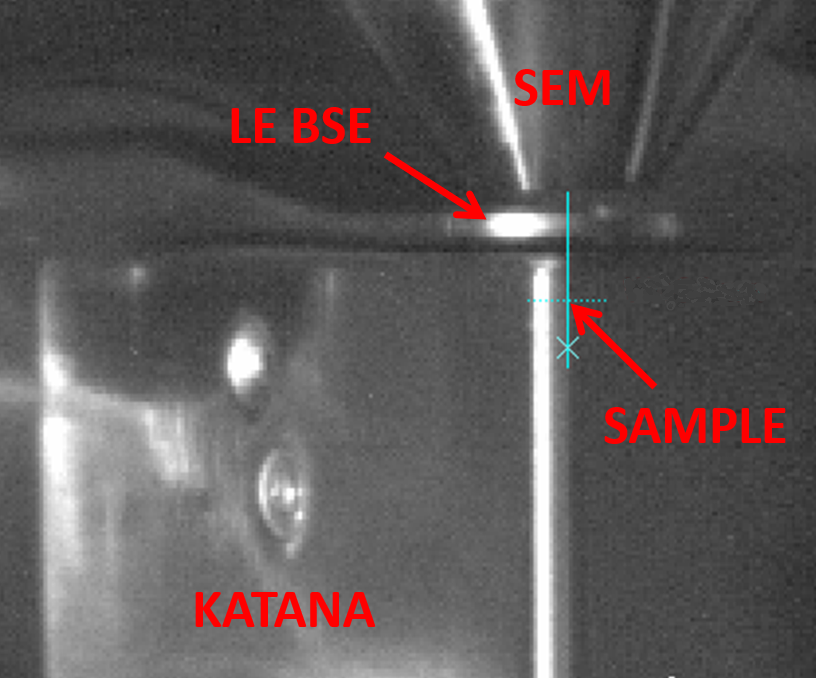
Figure 1. In-chamber Katana microtome mounted on a TESCAN stage.
Another integral part of SBF-SEM experiments is good quality back-scattered electron (BSE) signal detection. Therefore, we have developed a specialized low-energy BSE (LE BSE) detector with improved detection efficiency (particularly at low electron voltages and doses) and modified geometry to allow for imaging at low working distances (thus improving image resolution). During SBF imaging, the detector is inserted in between SEM column and Katana microtome with minimum spare gaps to decrease working distance to the sample inside the microtome – see Figure 2.
Figure 2. Spatial configuration of Katana microtome under SEM column with low-energy BSE detector in between.
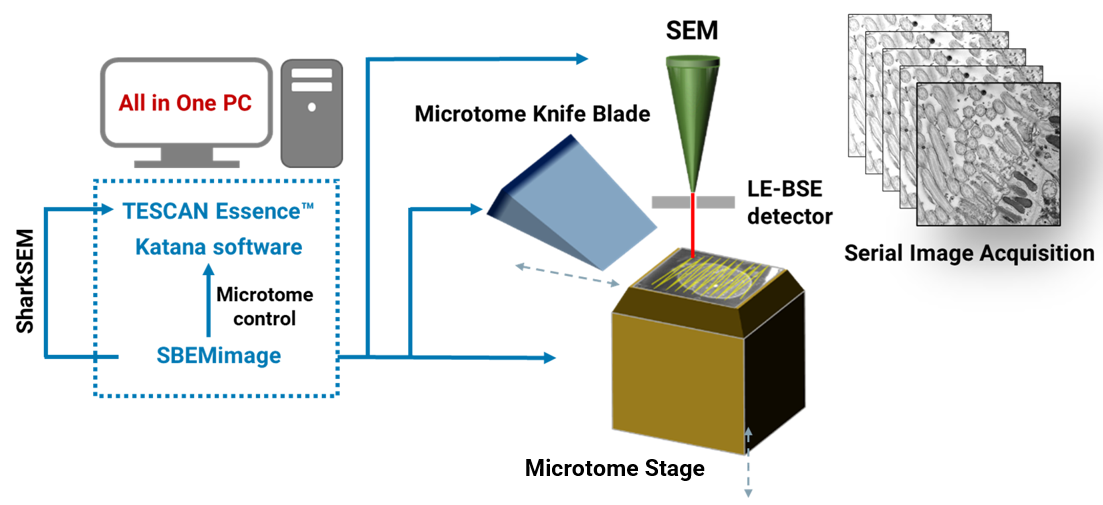
The automated execution of an actual SBF-SEM experiment is ensured by open-source Python-based acquisition software SBEMimage [4]. The application has been accommodated to communicate with TESCAN SEMs and ConnectomX microtome so it can coordinate SEM scanning and block-face cutting. SBEMimage provides users with outstanding flexibility in terms of imaging patterns and conditions, allowing for definition of complex tiled or multi-ROI acquisitions. It also features functions which are especially useful for long experiments, such as automatic SEM focusing, debris detection or remote monitoring with alerts. All SBF-SEM control software can be installed on a single computer to ensure smooth setup of the experiment (see Figure 3).
Figure 3. Schematic depiction of an automated SBF-SEM experiment using TESCAN SEM, ConnectomX Katana microtome and open-source acquisition software SBEMimage.
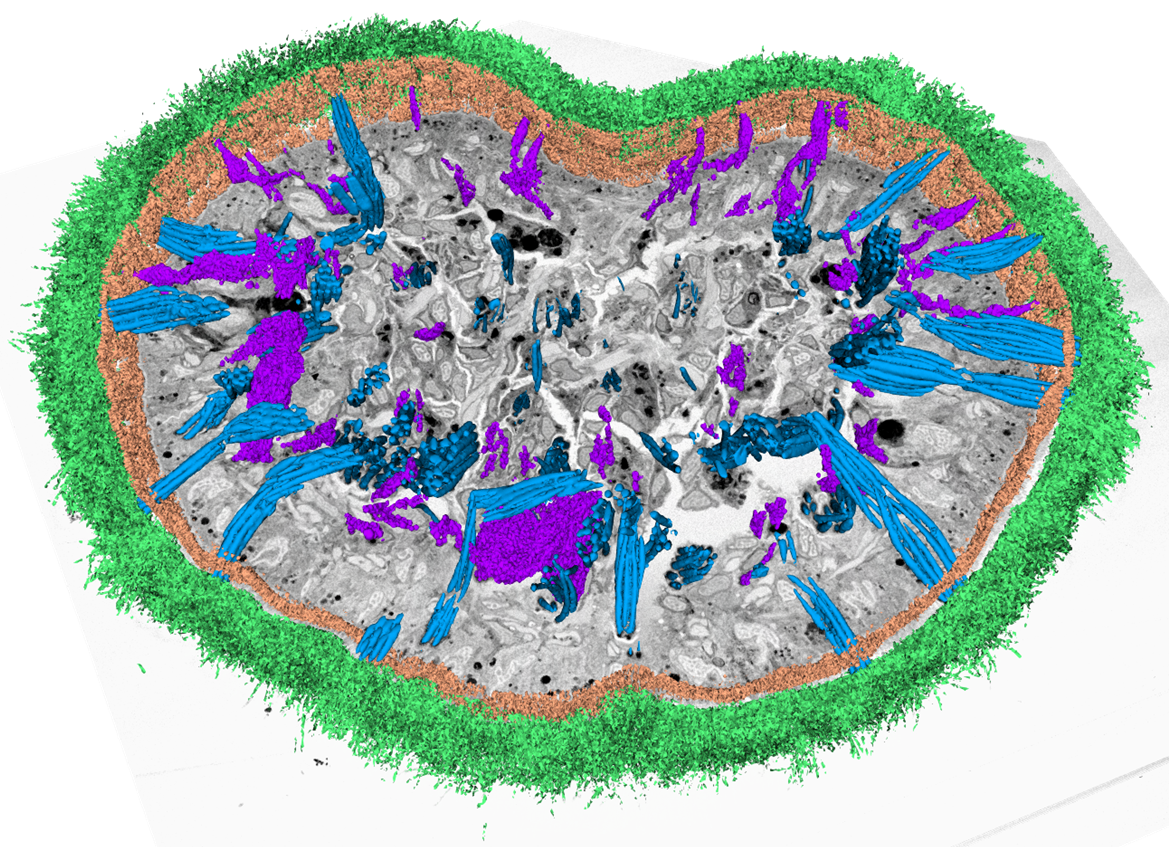
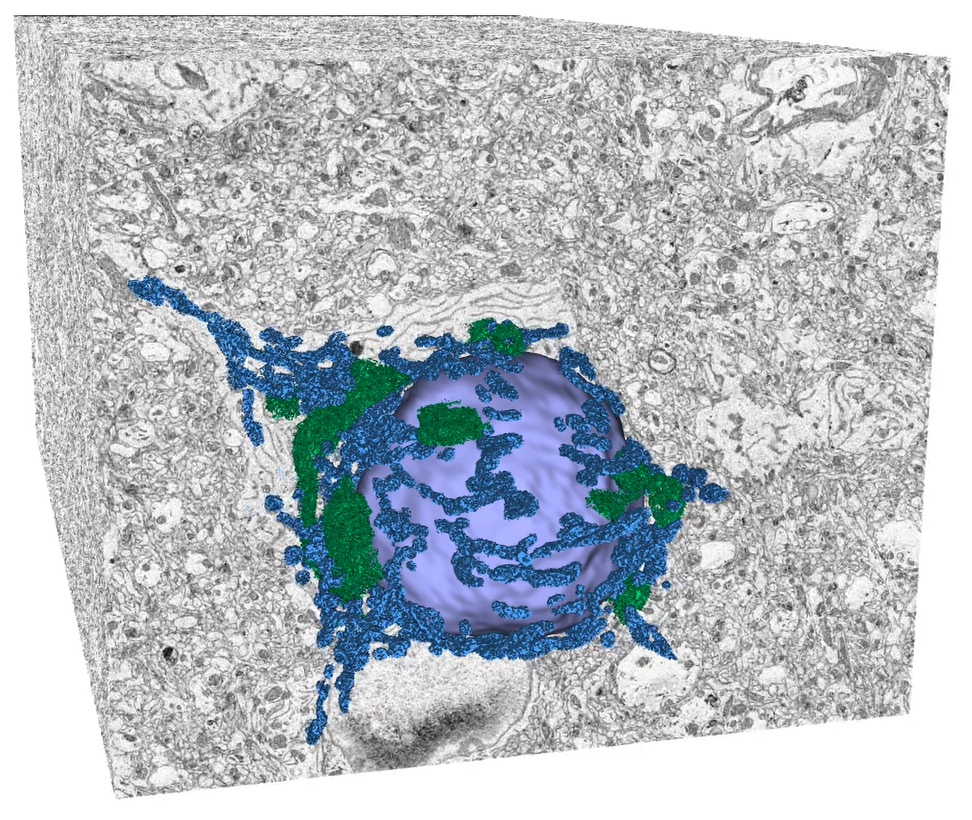
After SBF-SEM acquisition, multiple processing steps are necessary to transform raw data into scientifically relevant representation, which is usually a 3D volume visualization with subsequent segmentation and quantitative analysis. For these purposes, we have developed a specialized module in TESCAN 3D Analysis Suite (T3D), an in-house software for visualization of volume data. The software module offers functions optimized for all essential steps of volume EM data processing, from stitching of image tiles, detection and elimination of damaged slices through stack alignment and image enhancement methods to 3D reconstruction and visualization. With these software tools at hand, we are able to cover the entire SBF-SEM workflow in terms of data acquisition and processing (i.e. excluding sample preparation). An example of results achievable with TESCAN SBF-SEM solution can be seen in Figures 4 and 5, which showcase 3D analysis of flatworm and mouse brain samples, respectively.
Figure 4. 3D reconstruction and segmentation of Macrostomum lignano (flatworm) sample processed by TESCAN SBF-SEM workflow. Data acquired with TESCAN MIRA, data processing and visualization in T3D, segmentation in ORS Dragonfly. Sample courtesy of Dr. P. Ladurner and W. Salvenmoser, Department of Zoology, University of Innsbruck, Austria.
Figure 5. 3D reconstruction and segmentation of murine brain sample processed by TESCAN SBF-SEM workflow. Data acquired with TESCAN CLARA, data processing and visualization in T3D, segmentation in ORS Dragonfly. Sample courtesy of Stuart Searle, ConnectomX Ltd.
By providing integrated and compatible instruments, the presented SBF-SEM solution can help researchers achieve interpretable results faster, eliminating the need to repeatedly switch between different tools or to laboriously develop their own ad hoc workflows. Moreover, easy exchangeability of dedicated hardware components allows for quick reconfiguration of the SEM system, keeping microscope versatility intact. SBF-SEM has already proven its value and importance in biological research and we believe that these developments will make the technique even more accessible to a broad range of scientists.
Acknowledgments
The authors would like to thank Stuart Searle (ConnectomX Ltd.) and Benjamin Titze (SBEMimage creator and developer) for the collaboration on this project.
- References
[1] D. Smith and T. Starborg, Serial block face scanning electron microscopy in cell biology: Applications and technology, Tissue and Cell 57 (2019), p. 111-122. doi: 10.1016/j.tice.2018.08.011
[2] S. Lippens et al., Serial block face-scanning electron microscopy for volume electron microscopy, Methods Cell Biol. 152 (2019), p. 69-85. doi: 10.1016/bs.mcb.2019.04.002
[3] ConnectomX, https://www.connectomx.com (accessed Feb 02, 2023).
[4] SBEMimage, https://sbemimage.readthedocs.io (accessed Feb 10, 2023).