Direct detection camera as an alternative to negative staining for peptide structure determination
- Abstract number
- 238
- Presentation Form
- Poster & Flash Talk
- DOI
- 10.22443/rms.mmc2023.238
- Corresponding Email
- [email protected]
- Session
- Poster Session Two
- Authors
- Dr Julie A Watts (2, 6), Dr Cosimo Ligorio (1, 5), Professor Alvaro Mata (2, 4, 5), Dr Mike W Fay (3, 6)
- Affiliations
-
1. Biodiscovery Institute, University of Nottingham
2. School of Pharmacy, University of Nottingham
3. School of Physics, University of Nottingham
4. Biodiscovery Institute University of Nottingham
5. Department of Chemical and Environmental Engineering, University of Nottingham
6. Nanoscale and Microscale Research Centre, University of Nottingham
- Keywords
Transmission electron microscopy
Peptides
nanofibres
direct electron detection
negative staining
low dose
- Abstract text
Peptides may be regarded as molecular building blocks for formation of cell-instructive biomaterials, self-assembling into nanofibers and nanofibrillar meshes. Facile imaging of peptide structures is essential to optimize formulations of materials with enhanced properties. Transmission electron microscopy (TEM) of peptide assemblies traditionally involve use of heavy metal stains, most commonly Uranyl Acetate (UA), to confer electron density, but UA artifacts are often observed. UA is toxic and radioactive, with cumulative exposure, and is an environmentally hazardous substance. Therefore, reducing use would be advantageous.
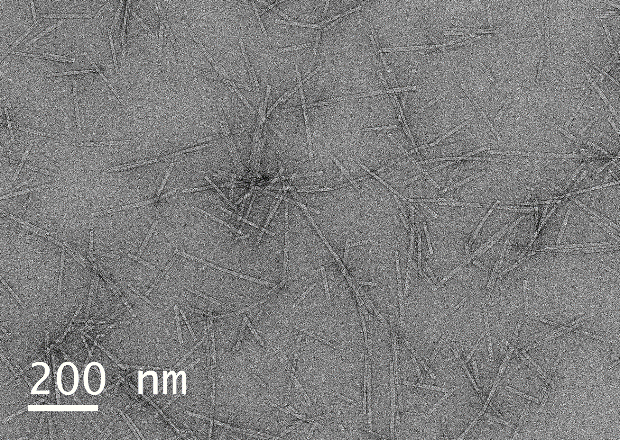
Here we present images acquired using a single-electron direct detection camera from samples that were not UA stained, showing clear evidence of peptide structures. For comparison the same samples were UA stained and micrographs recorded under the same conditions. Peptides in lyophilised form (0.01 mg/ml in 10 mM HEPES) were drop cast onto a carbon-coated copper grid, with and without the addition of UA. Samples were observed in a JEOL 2100F TEM, operating at 200kV, and a Gatan K3 IS direct electron camera in counting mode. Images were recorded at an electron dose rate of ~1 or 3 e-/Å2/s, for unstained and stained samples, respectively, in both cases 64 frames were summed (0.03s per frame). Images were post-processed by a python script utilising the non-linear de-noising means filter from the scikit-image package, and measured by calculating the visible thickness from a 100 pixel averaged line profile in Gatan Microscopy Suite. Samples not UA stained showed clear nanofibre structures, with a size distribution of 18.6±3.9nm and some assembly of cylindrical micelles in fibrillar meshes (Figure 1a). By comparison, samples prepared by UA staining also showed nanofibre structures with a comparable size distribution of 19.9±1.1nm. (Figure 1b). We also note the presence of individual cylindrical micelle structures in stained micrographs, which suggests that UA stabilises the peptide nanofibers, in addition to providing contrast. There is no significant size difference observed, and direct evidence of peptide nanofibers without UA staining artefact-free and obtained without the need for toxic, radioactive UA stain.
a
b
Figure 1 (a) Unprocessed images of Peptide self-assemblies unstained, (b) Peptide self-assemblies prepared using traditional UA staining.