Label-Free High Content Screening for Phenotypic Drug Discovery
- Abstract number
- 557
- Presentation Form
- Poster
- DOI
- 10.22443/rms.mmc2023.557
- Corresponding Email
- [email protected]
- Session
- Poster Session Three
- Authors
- Dr Peter Djali (1), Dr Meetal Solanki (1)
- Affiliations
-
1. Phasefocus
- Keywords
HCS, High Content Screening, Drug Discovery, Pheneptypic Screening, Label-free
- Abstract text
Phenotypic screening in drug discovery is now commonplace. Early high-content imaging systems paved the way as they revealed feature specific phenotypes based on simple labelling techniques. One of the first widely used phenotypic screens was nuclear factor kappa B (NFkB) nuclear translocation and its automated analysis. Wider uptake, an increased range of dyes and labels, improved processing algorithms and computer power make it now commonplace to perform phenotypic screens with cells “painted” with a whole array of dyes.1 These can, for example, be used to study the mechanism of action (MOA), toxicity and many other parameters. However, there are still limitations with the use of cell painting. The dyes are predefined and measure what they are designed to, removing the chance for serendipitous observations that can be discovered in a more open screen. Painting is expensive and the cost per well cannot easily be justified. The use of multiple dyes and the resulting susceptibility to phototoxicity means by definition it is not applicable to live-imaging and must be used in an end point assay. In turn, the process of dye fixing or stabilisation for one dye may adversely affect the performance of another.
Kinetic screening of live cells can therefore present an alternative, but until now the limitations of what can be measured in live cell screens are considerable and based on the toxicity or phototoxicity of common dyes with limited information available from a conventional phase contrast image. Many primary cells are too sensitive to long term illumination for even green emitting proteins and many other dyes will perturb long term cell behaviour in one way or another e.g. DNA intercalating dyes are highly disruptive to normal cell division. The perfect solution is to get as much data in an unlabelled format as possible, whilst combining this with as few phototoxic dyes as possible.
Here we present how completely label-free Quantitative Phase Imaging with the Livecyte from Phasefocus, can provide insights into live cell behaviour that cannot be reliably measured by other techniques. We also show how a common dye described as suitable for live cell imaging perturbs cell behaviour.
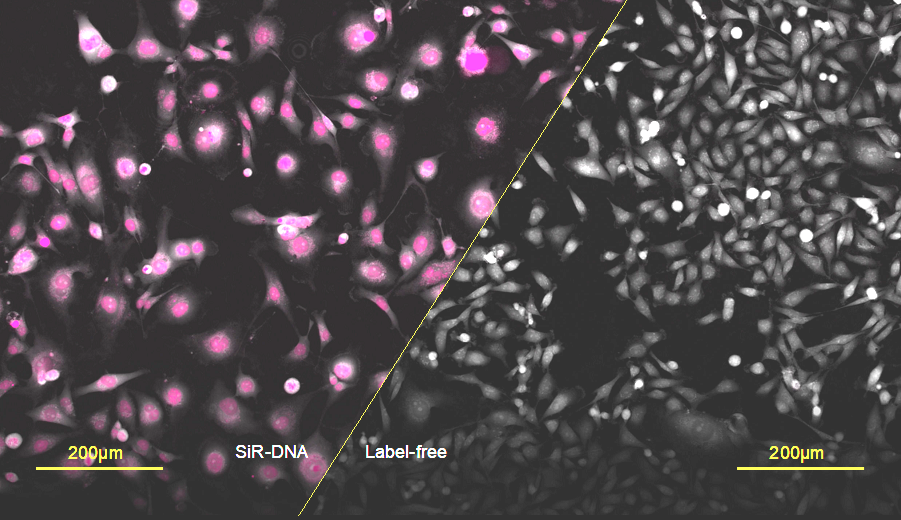
Figure 1: An illustration depicting label-free imaging versus SiR-DNA fluorescence. MDA-MB-231 cells were exposed to 250nM SiR-DNA at 400µW/mm2 for 120 hours. Fluorescent treated MDA-mb-231 cells appear to be larger with a less characteristic epithelial phenotype compared to unlabelled cells
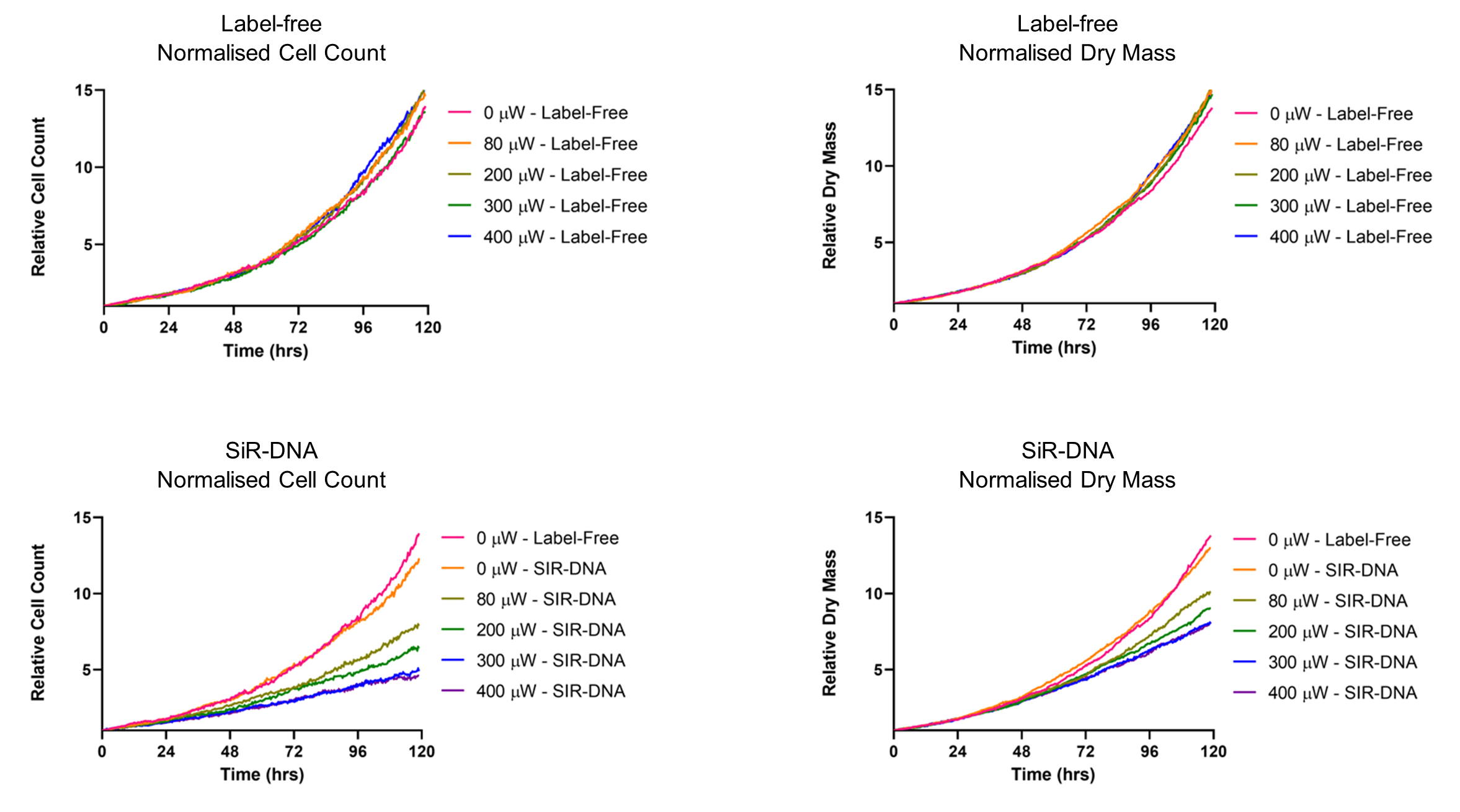
Figure 2: Illustrative graphs showing the relative cell count over time for unlabelled (a) and SiR-DNA labelled cells (b) at varying LED powers. SiR-DNA labelling combined with illumination inhibits cell proliferation. Relative Dry Mass over time for unlabelled (c) and SiR-DNA labelled cells (d) at varying LED powers SiR-DNA labelling combined with illumination inhibits the overall accumulation of dry mass per field of view.
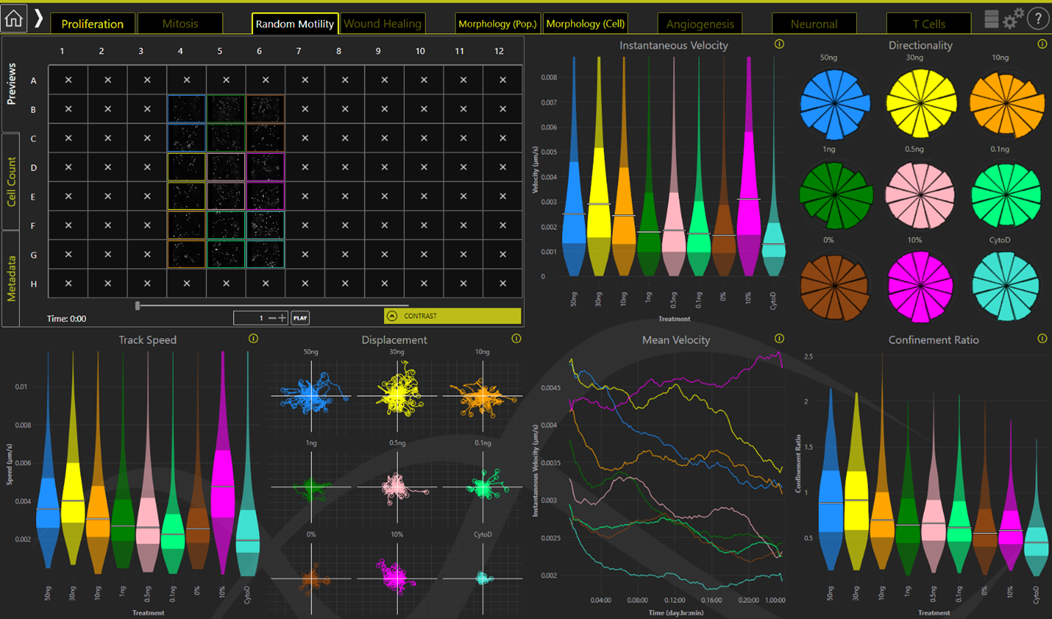
Figure 3: Illustrative graphs showing Motility Dashboard on Livecyte’s Analyse software. Livecyte analysis solution is particularly designed for kinetic analysis. It tracks and segments every cell leading to a plethora of phenotypic outputs including Random Motility (pictured), Proliferation, and Morphology. These are displayed in are easy to use dashboard format.
- References
1 Bray, M.A., Singh, S., Han, H., Davis, C.T., Borgeson, B., Hartland, C., Kost-Alimova, M., Gustafsdottir, S.M., Gibson, C.C. and Carpenter, A.E., 2016. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nature protocols, 11(9), pp.1757-1774.