Compressive Cryo FIB-SEM Tomography
- Abstract number
- 161
- Presentation Form
- Contributed Talk
- DOI
- 10.22443/rms.mmc2023.161
- Corresponding Email
- [email protected]
- Session
- EMAG - 3D & Tomographic Electron Microscopy
- Authors
- Daniel Nicholls (3), Jack Weels (4), Alex Robinson (3), Amirafshar Moshtaghpour (3, 2), Maryna Kobylynska (1), Roland Fleck (1), Angus Kirkland (2, 5), Layla Mehdi (3, 6, 7), Nigel Browning (3, 6, 7, 8)
- Affiliations
-
1. Centre for Ultrastructural Imaging, King’s College London
2. Correlated Imaging Group, Rosalind Franklin Institute, Harwell Science and Innovation Campus
3. Department of Mechanical, Materials and Aerospace Engineering, University of Liverpool
4. Distributed Algorithms Centre for Doctoral Training, University of Liverpool
5. Department of Materials, University of Oxford
6. Physical and Computational Science Directorate, Pacific Northwest National Laboratory,
7. The Faraday Institution, Quad One, Harwell Science and Innovation Campus
8. Sivananthan Laboratories
- Abstract text
Cryo focused ion beam scanning electron microscope (FIB-SEM) tomography allows 3-dimensional sections of biological materials to be serially milled and imaged at the nanoscale [1]. These experiments are however limited by time, both in the removal of material and in imaging, and by damage due to the inherently destructive nature of both the ion and electron beams. Compressive sensing (CS) [2,3] and a novel targeted sampling method [4] are proposed to be valuable tools in overcoming these limitations of cryo FIB-SEM tomography and a simulated experiment is presented here to test the validity of the method in principle.
Compressive sensing is a method of forming incomplete optimally acquired data followed by a form of data reconstruction to allow the data to be analysed. CS has seen positive applications to scanning electron microscopy and scanning transmission electron microscopy in recent years, in particularly through the application of probe subsampling; the manipulation of the electron beam scanning coils to follow a sampling pattern, rather than a traditional space-filling raster scan. The data is then typically recovered using an inpainting algorithm, of which many exist.
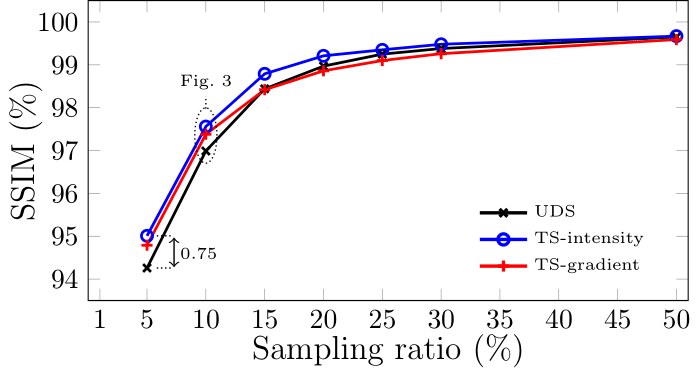
In cryo FIB-SEM tomography, a focus ion beam is used to remove a layer of a material and a focused electron beam is then used to image the newly revealed surface, with the process being repeated as many times as required. The proposed targeted sampling methods aim to take advantage of the layer-to-layer self-similarity of the successive slices by designing the sampling pattern for the next slice based on the previously acquired data. The specific targeted sampling methods used here are ‘TS-intensity’ and ‘TS-gradient’, wherein the sampling pattern is probabilistically determined by the intensity and gradient, respectively, of the previous acquired image (with ‘TS’ meaning targeted sampling). The results for this are presented in Fig. 1, where these targeted sampling methods are compared to uniform density sampling (UDS), often called ‘random sampling’. For this study, a cryo FIB-SEM tomography data cube of a Euglena gracilis (E. gracilis) cell was acquired at KCL and the aforementioned methods applied in silico. These methods provided an overall increase in reconstructed image quality, as determined by the calculated structural similarity index measure (SSIM), at all sampling percentages tested and were particularly effective at low sampling rates, where the most benefit of CS can be gained. Several examples images are shown in Fig. 2 at various points throughout the data cube for the three methods tested with magnified insets to aid visual validation. Note that drift correction has not been applied in this application, though it is theorised to prove beneficial.
Figure 1. Performance comparison between the two proposed sampling strategies and UDS over a range of sampling ratios.
Figure 2. Examples of treated data from E. gracilis data cube. The ground truth and corresponding reconstructed images from 10% of the measurements are shown. Visually, all three methods of subsampling provide good quality reconstructions. Targeted sampling provides slightly increased sharpness and spatial fidelity.
- References
[1] R. A. Fleck et al in “Biological Field Emission Scanning Electron Microscopy” (2019), John Wiley & Sons.
[2] D. L. Donoho, IEEE transactions on information theory, vol. 52 (2006), no. 4, pp. 1289–1306
[3] E. J. Candes et al, IEEE transactions on information theory, vol. 52 (2006), no. 2, pp. 489–509.
[4] D. Nicholls et al, arXiv preprint (2022), arXiv:2211.03494