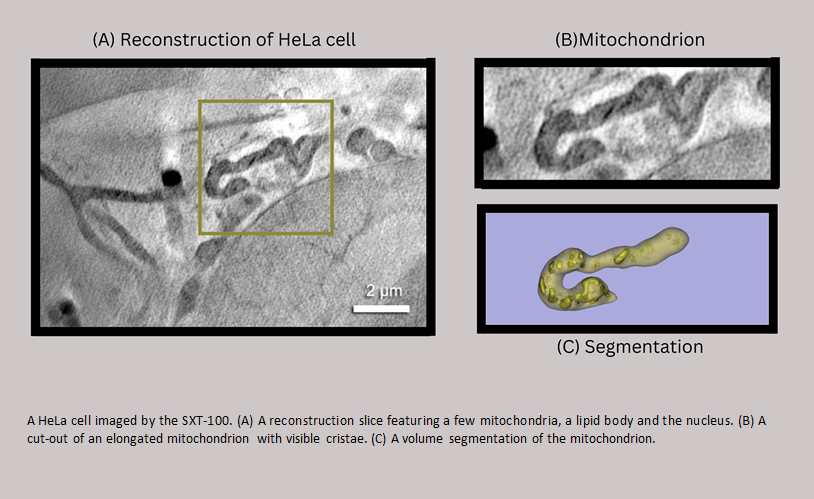
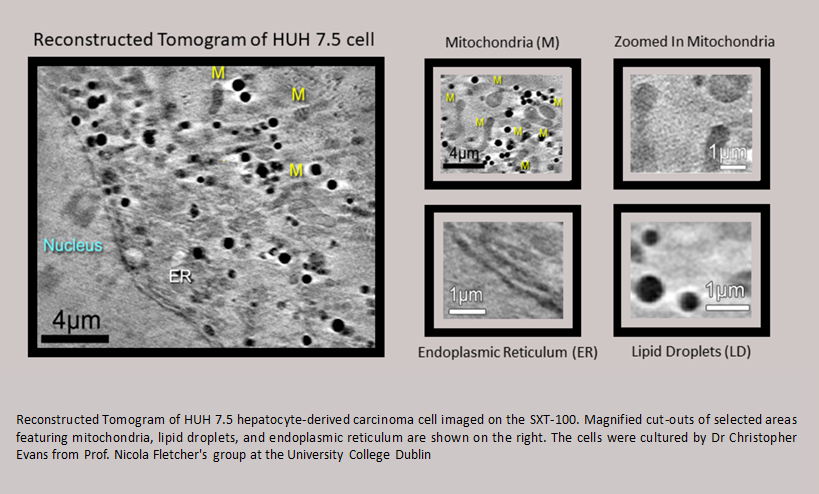
Laboratory-based cryo-Soft X-ray Tomography
- Abstract number
- 123
- Presentation Form
- Contributed Talk
- DOI
- 10.22443/rms.mmc2023.123
- Corresponding Email
- kenneth.fahy@siriusxt.com
- Session
- Correlative and Multimodal X-ray Microscopy
- Authors
- Kenneth Fahy (1), Paul Sheridan (1), Sergey Kapishnikov (1), William Fyans (1), Fergal O'Reilly (1, 2), Tony McEnroe (1)
- Affiliations
-
1. SiriusXT
2. UCD Dublin
- Keywords
Soft x-ray tomography, 3D cell structure, whole cell imaging, label-free imaging, disease research, viral infection, anti-viral drugs
- Abstract text
Analysis of three-dimensional biological cell samples is critical for understanding the mechanisms of viral disease and for developing novel therapeutics. Soft X-ray microscopy (SXM), employing low energy X-rays (E ~500 eV) in the so called ‘water-window’, is the unique technology that can image whole intact cells in 3D under normal and pathological conditions without labelling or fixation, at high throughput and spatial resolution [1-4]. The main challenge of SXM is that the photonic illumination required for imaging has heretofore only been available at synchrotron labs [5].
SiriusXT has developed a compact soft X-ray microscope (the “SXT-100”) for three-dimensional imaging of whole cells and tissue sections that can be performed in a laboratory. The capabilities of this imaging device, combined with complementary light and electron microscopy approaches, is currently being demonstrated through a series of virology Use Cases to generate new scientific knowledge on the viral life cycle and host cell response to viral infection (EU project “CoCID”) [6-8].
Our studies demonstrate the benefits of the lab-based system relative to both synchrotron based SXM as well as other imaging modalities. However, the most comprehensive view of the complex structure of cells is unlikely to come from a single microscope. Integration of SXM into CLEM imaging workflows offers a powerful new method for 3D imaging of whole cells and tissue. While SXM quantitatively and rapidly images fully hydrated, intact cells at about 50 nm resolution, cryo-TEM generates high-resolution (~5 nm) data from small regions of interest of cells. Combining these two powerful imaging techniques with data from fluorescence microscopy will provide a more informative picture.
We can facilitate correlation of SXM with light and electron microscopy by integration of a fluorescence microscope, dual modality of sample presenting scheme, including EM grids and FEI-Autogrids. We are also developing ultra-thin-walled glass capillaries as a sample holder to facilitate full tilt tomography.
Here we present tomography data from whole vitrified cells (healthy and infected) and discuss the development of novel correlative workflows that can benefit from soft X-ray imaging.
- References
References
[1] M Harkiolaki et al., Emerging Topics in Life Science 2 (2018), p. 18. doi: 10.1042/ETLS20170086
[2] J Guo and CA Larabell, Current Opinion in Structural Biology 58 (2019), p. 324. doi: 10.1016/J.SBI.2019.06.012
[3] G Schneider et al., Nature Methods 7 (2010), p. 985. doi: 10.1038/NMETH.1533
[4] FJ Chichón et al., Journal of Structural Biology 177 (2012), p. 202. doi:10.1016/J.JSB.2011.12.001
[5] A Ekman et al., Synchrotron Light Sources and Free-Electron Lasers Springer, Cham. (2019) https://doi.org/10.1007/978-3-319-04507-8_43-3
[6] K Fahy et al., JPhys Photonics 3 (2021), doi: https://doi.org/10.1088/2515-7647/abfc5a
[7] https://cocid.eu/
[8] The authors acknowledge funding from the European Union’s Horizon 2020 Research and Innovation programme (No. 959776, project LiCENT and No. 101017116, project CoCID) as well as the Irish Research Council (No. EBPPG-2020-278).