Correlative cryo-bioimaging to study coronavirus replication organelles
- Abstract number
- 76
- Presentation Form
- Poster & Flash Talk
- DOI
- 10.22443/rms.mmc2023.76
- Corresponding Email
- [email protected]
- Session
- Public Health: The Impact of Microscopy
- Authors
- Mr Patrick Phillips (1, 3, 2), Dr Helena Maier (3), Prof Philippa Hawes (3), Dr Maria Harkiolaki (1), Dr Dan Clare (1), Prof Jonathan Grimes (2)
- Affiliations
-
1. Diamond Light Source
2. Oxford University
3. The Pirbright Institute
- Keywords
Correlative imaging, fluorescence, cryo, soft X-ray tomography, electron tomography, coronavirus, replication organelle
- Abstract text
The Gammacoronavirus Infectious Bronchitis Virus (IBV) is a highly contagious pathogen of poultry and can be used as a model coronavirus (CoV) to study the formation and structure of CoV replication organelles (ROs). ROs are platforms for the viral RNA synthesis machinery and form by rearranging host-cell membranes. A variety of conserved RO structures exist including double membrane vesicles (DMVs) and double membrane spherules (DMSs). The mechanism of DMV formation and the function of DMSs have not yet been elucidated. Using fluorescent recombinant viruses and correlative cryo-bioimaging, sites of RO membranes will be marked and directly targeted for imaging using X-rays and electrons. These techniques will reveal RO structures in 3D, to high resolution, and under near-native conditions.
Fluorescent tags were inserted at the N-terminus of nonstructural protein 2 (nsp2) using reverse genetics. Recombinant IBV growth kinetics were characterised. Immunofluorescence imaging was used to confirm that fluorescent-nsp2 signal correlated to sites of nascent viral RNA synthesis (Figure 1). Resin-embedded correlative light and electron microscopy (CLEM) proved that fluorescence signal from tagged-nsp2 marks known IBV RO structures (Figure 2).
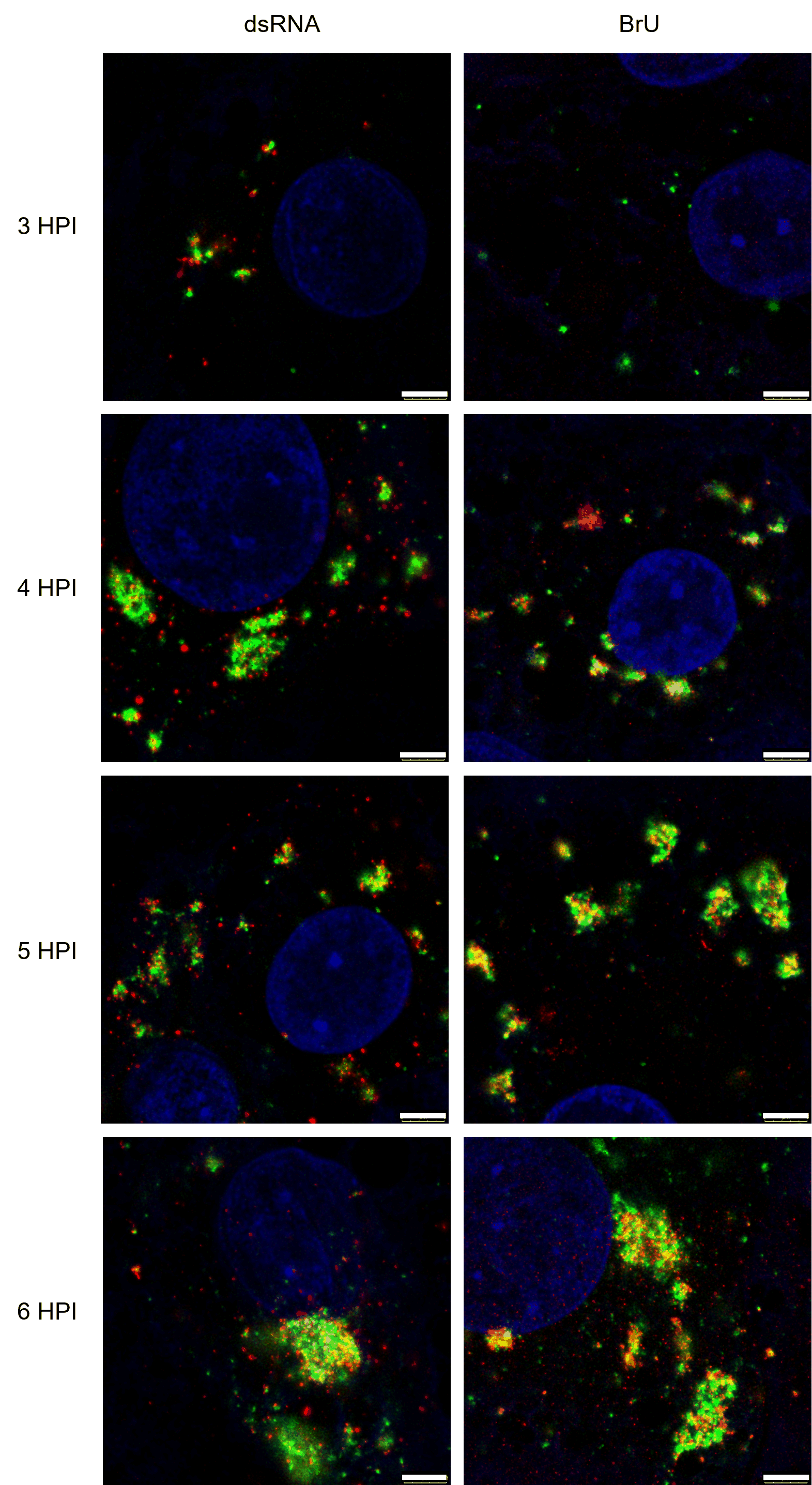
Figure 1. STED images of BeauR-eGFP-nsp2 (green) colocalising with dsRNA (red, left column) and nascent viral RNA (BrU, red, right column). CK cells were infected with BeauR-eGFP-nsp2 (green), cells were fixed at the specified times and labelled with anti-dsRNA (left; red) or anti-BrU (right; red). For BrU labelling, cells were treated with BrU and ActD 30 minutes prior to fixation and kept in an RNase-free environment. Nuclei (blue) labelled with Topro3. Scale bars are 3 µm.
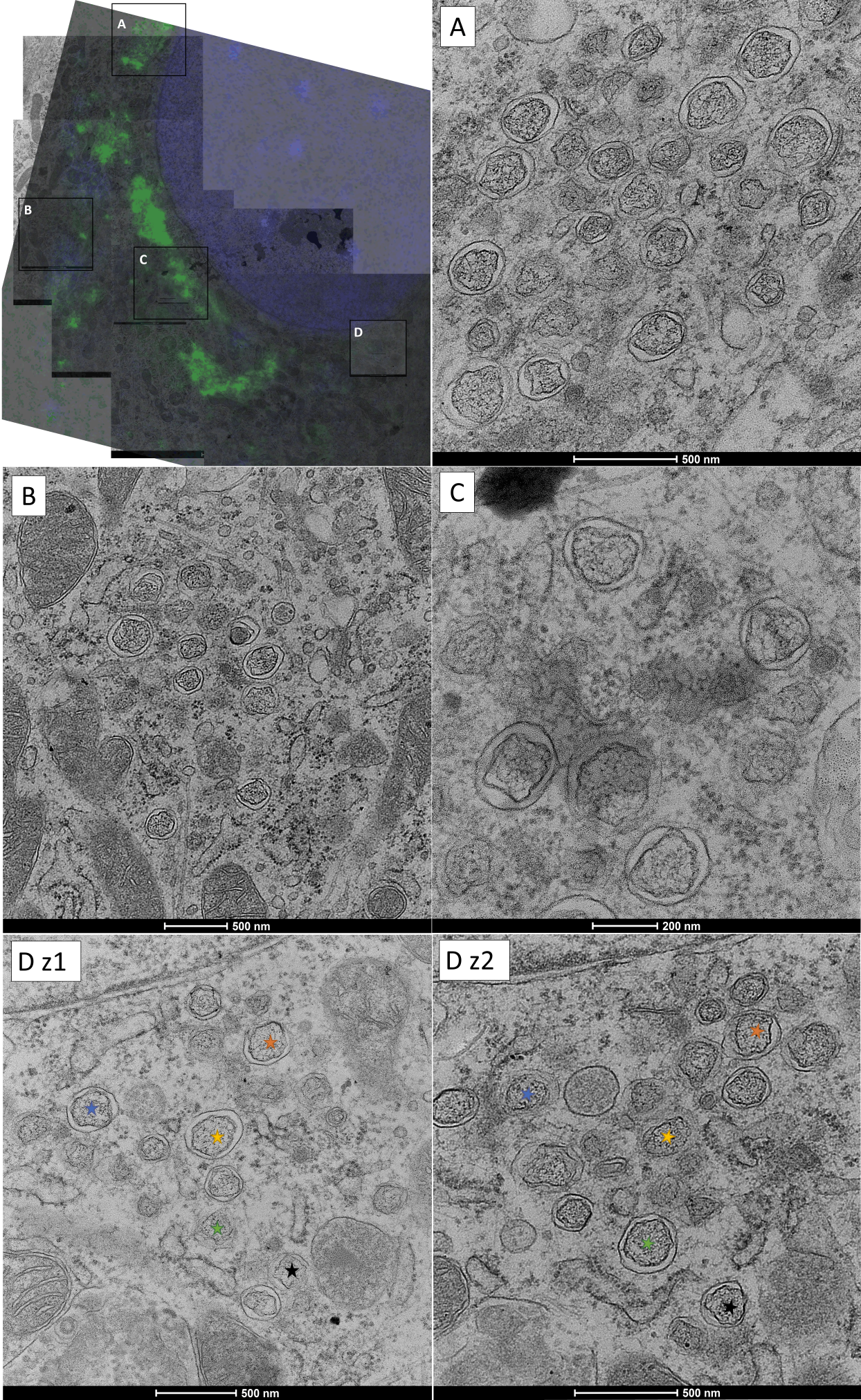
Figure 2. Correlative light and electron microscopy (CLEM) of BeauR-eGFP-nsp2 infected CK cell at 6hpi reveals GFP-nsp2 signal marks infected cells and can direct imaging to specific RO membrane patches within the cell cytoplasm. CK cells were infected with BeauR-eGFP-nsp2 (green), nuclei were labelled with Hoechst (blue) and live cells were imaged to identify cells with eGFP-nsp2 signal. Fluorescence information was captured before fixing cells at 4-6 hpi, staining, dehydration, resin embedding and sectioning. The pattern of the nuclei from the live cell fluorescence images is correlated to the pattern of nuclei on the electron micrographs to identify the infected cell for which the patterns of eGFP-nsp2 signal were captured. Electron micrographs of this cell were then collected at higher magnification to observe the RO structures. The same cell can be found in other sections on the grid to give information from different z-depths. Fluorescence data and the electron micrographs can then be overlayed to see the membranes which correlate to the eGFP-nsp2 signal. Darkest membranes represent zER and DMSs, dark rings represent DMVs. Nuclei (blue) labelled with Hoechst. The top left panel shows the correlation between fluorescence and electron microscopy with RO patches in boxes A-D. Patrial DMSs and zER can be seen in C. The DMV patch captured in box D was observed in two separate microtomy slices (sections) of the same sample 160nm apart in z, the same DMV in each z-slice is labelled with the same-coloured star.
At B24 (Diamond), structured illumination microscopy (SIM), for super-resolution fluorescence localisation of early RO membrane rearrangements, and cryo-soft X-ray tomography (cryo-SXT), for natural contrast imaging of ROs throughout the depth of whole cells to 25nm resolution, is being used to reveal forming DMVs and RO interactions throughout whole cells. Also, at eBIC (Diamond), fluorescence-guided focused ion beam milling and scanning electron microscopy (FIB-SEM) and cryo-electron tomography (cryo-ET; with subtomogram averaging) is being used to observe DMSs. DMSs were first seen in IBV and high resolution, native-state imaging is expected to identify their associated proteins which may give clues to their function (Maier et al., 2013). This work will contribute to understanding the sites of coronavirus RNA synthesis and this critical stage of the virus lifecycle.
- References
Maier, H. J., Hawes, P. C., Cottam, E. M., Mantell, J., Verkade, P., Monaghan, P., Wileman, T., & Britton, P. (2013). Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio, 4(5), e00801–e813. https://doi.org/10.1128/mBio.00801-13