FLIMbow: a multicolor multi-lifetime labeling method for individual cells and cell lineages
- Abstract number
- 322
- Presentation Form
- Poster Flash Talk + Poster
- DOI
- 10.22443/rms.mmc2021.322
- Corresponding Email
- [email protected]
- Session
- Stream 5: Imaging in Development and Disease
- Authors
- Vasilisa Polinovskaya (3), Igor Kramarev (3), Dmitry Gorbachev (3), Ilya Solovyev (1), Alexander Savitsky (1), Maria Lukina (2), Lyubov Shimolina (2), Vadim Elagin (2), Marina Shirmanova (2), Konstantin Lukyanov (3)
- Affiliations
-
1. A.N. Bach Institute of Biochemistry, Research Center of Biotechnology of the Russian Academy of Sciences
2. Privolzhsky Research Medical University
3. Skolkovo Institute of Science and Technology
- Keywords
FLIM, Brainbow, multiparameter imaging, cell lineage
- Abstract text
Brainbow and the related multicolor cell-labeling methods is an elegant strategy that has found numerous applications in the following research areas: cell lineages tracking in embryology and stem cell biology, connectome mapping in neuroscience, monitoring of tumor growth in cancer biology, and so on [1]–[3]. Brainbow allows for permanent markings (labeling) of target cells in multiple distinct colors by combining and expressing 3-4 fluorescent proteins (FPs) at different ratios in each cell. In general, Brainbow with three FPs can generate a palette reaching up to 100 colors. However, it is almost impossible to distinguish between the close hues and increase their due to natural experimental especially in in vivo research. Moreover, each FP occupies the corresponding fluorescence channel, leaving no spectral space for marking any objects of interest.
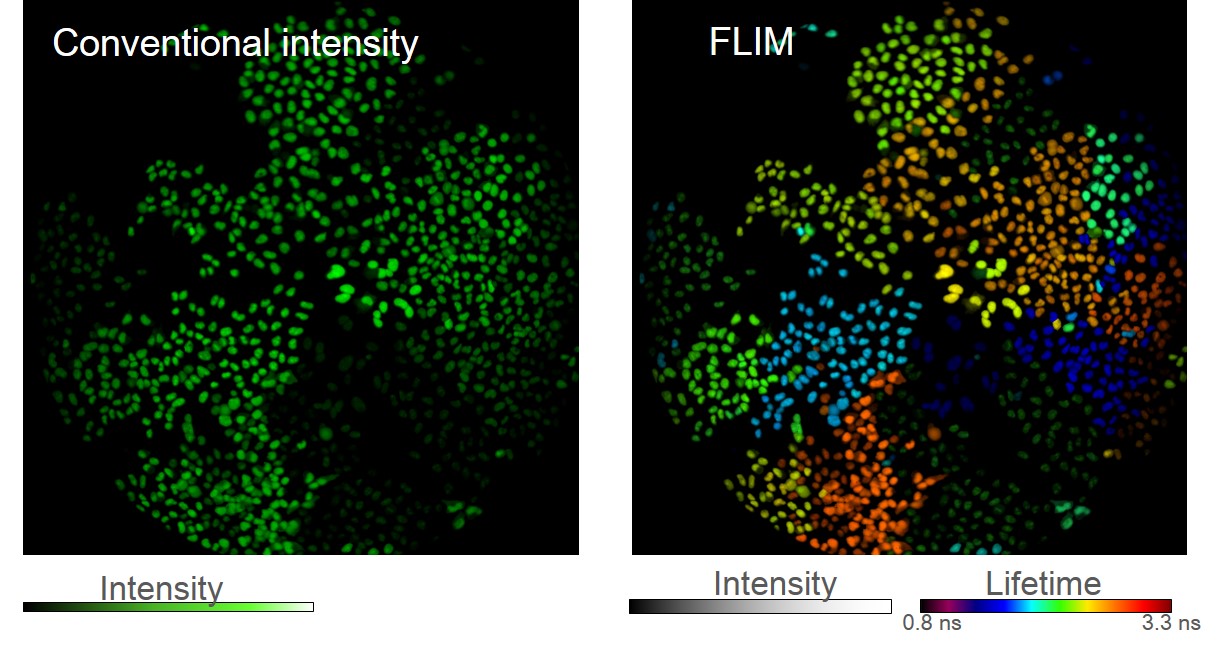
We developed the FLIMbow method, which expanded the palette of labels in a new dimension, using two independent parameters to distinguish objects: not only the color of the fluorescence but also the lifetime. We visualized in the Fluorescence Lifetime Imaging Microscopy (FLIM) mode cells stochastically expressing FPs of a similar spectrum, but with different lifetimes. We have revealed that these cells have distinguishable pseudo colors, similar to the effect in spectral Brainbow, but only within one fluorescent channel. We have shown that lentiviral vector-mediated multi-lifetime marking remained stable after cell division, thus allowing the analysis of clonal cell fates in vitro and in vivo (Fig.1). Expansion of the pool of fluorescent proteins into two extra fluorescent channels increases the amount of information about each labeled cell, which in turn fine-tunes the accuracy of determining the cell to a particular clone. As proof of principle, we studied tumor development in mice. FLIMbow multi-parameter marking technology has shown both quantitative and qualitative superiority over the original and related Brainbow methods.
Figure 1. Hela cells labeled with green FLIMbow lentiviruses, сell clones retain their pseudo-colored labels during division.
- References
[1] J. Livet et al., “Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system,” Nature, vol. 450, no. 7166, pp. 56–62, 2007, doi: 10.1038/nature06293.
[2] K. Weber et al., “RGB marking facilitates multicolor clonal cell tracking ” Nat. Med., vol. 17, no. 4, pp. 504–509, 2011, doi: 10.1038/nm.2338.
[3] S. Lamprecht et al., “Multicolor lineage tracing reveals clonal architecture and dynamics in colon cancer,” Nat. Commun., vol. 8, no. 1, pp. 1–8, 2017, doi: 10.1038/s41467-017-00976-9.