High precision autofocus on a low cost microscope: automating blood sample imaging on the OpenFlexure Microscope
- Abstract number
- 127
- Presentation Form
- Submitted Talk
- DOI
- 10.22443/rms.mmc2021.127
- Corresponding Email
- [email protected]
- Session
- Stream 2: Software and Smart Microscopy
- Authors
- Mr Joe Knapper (2), Dr Joel Collins (2), Dr Julian Stirling (2), Dr Samuel McDermott (3), Dr William Wadsworth (2), Dr Catherine Mkindi (1), Dr Richard Bowman (2)
- Affiliations
-
1. Ifakara Health Institute
2. University of Bath
3. University of Cambridge
- Keywords
focus, sharpness, microscopy, diagnosis, malaria, open-source
- Abstract text
The OpenFlexure Microscope (OFM) is a low-cost, 3D printed microscope which can be customised for a range of uses [1]. These uses – including academic research, outreach, and education – each require different budgets, imaging modes and components. The highest quality OFM costs around £200 in parts, and includes a Raspberry Pi, Pi Camera and stepper motors. The motors control the microscope translation stage, allowing the user to automatically capture images across a sample. The OpenFlexure software allows the user to select the range of movement (up to 12×12×4mm) and distance between images in x, y and z. Due to the geometry of the stage, movements in x and y (in the sample plane) are not truly orthogonal, leading to slight drift in z. This drift is especially noticeable in high-resolution imaging, in which the smaller depth of field means that even slight drift can cause the images to become defocused. Microscopists can correct for this manually by refocusing at each xy location, but for fully automated microscopy, a reliable autofocus procedure is required. We compare different approaches of measuring image sharpness and positioning the microscope, ensuring that all solutions are suitable for the design and resources of the OFM.
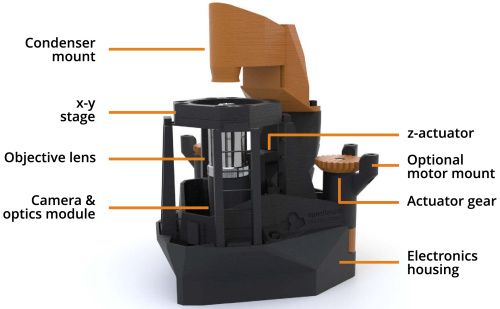
Figure 1: A labelled render of the OpenFlexure Microscope
The automated image collection enabled by the OFM has several benefits over manual microscopy. The images can be saved and tiled to create a full representation of the sample. It also improves the user’s workflow, automating data acquisition which allows users to work on other tasks. As a result, the OpenFlexure Microscope is being developed to support the diagnosis of malaria in sub-Saharan Africa. Manual optical microscopy is described by the World Health Organisation as the “gold standard” of malaria diagnosis, requiring a technician, Giemsa-stained blood sample, and high-magnification microscope. Automating the image collection would improve the throughput of the clinics, but requires a precise autofocus procedure, as the 100× objectives used have a depth of field of ~1 μm.
OpenFlexure software includes two focus metrics [2], each of which award a higher score to images with sharp features and hard edges. The first is a modified 2D Laplacian, which applies a convolution to a greyscale image [3]. This convolution awards each pixel a score reflecting the rate of change around it, which is then raised to the fourth power and averaged to give a sharpness score. This method has been found to be reliable but slow, and is heavily affected by noise. The second metric utilises the software’s MJPEG stream, which is included to give the user a preview of the microscope field of view. In JPEG encoding, each image is divided into small blocks, which are described as a superposition of discrete cosine functions. The encoding uses the fewest discrete cosine functions possible, meaning that sharper images require more functions and therefore more storage space. This method of identifying the sharpest image is quick and computationally ‘free’, as the MJPEG stream is used by default in the software. However, it can fail on sparse samples as small, out of focus objects may overflow into otherwise empty blocks, increasing the image size. Blood samples are densely populated, and so this limitation is accepted.
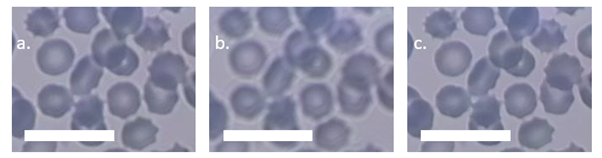
Figure 2: Cropped blood sample images from the OFM of the same xy position using different autofocus procedures. a) is an image taken after performing an MJPEG autofocus. b) is the centre image from a 9-image z-stack centred on the same z-position as a). The defocusing in b) is a result of the positional uncertainty introduced by performing the z-stack. c) is the sharpest image from a 9-image rolling z-stack, which accounts for backlash and uses fitting to determine if the sharpness is sufficiently high and centred within the stack. Scale bars are 20 μm.
The OpenFlexure Microscope is currently being used to scan blood samples in Tanzanian health institutes, taking 900 images per sample in a 10×10×9 grid. The stack of 9 images, each separated by 0.5 μm in z, is centred on the estimated focal point of each xy location, using the MJPEG method over a range of 100 μm. The central image from each stack are tiled together to produce a large recreation of the blood sample, while the remaining images are used to assess the focus of the stack. The effect of backlash on a stack of images separated by approximately 4 μm is significant; as the microscope changes direction at the start of the stack, the gaps between the teeth of the gears moving the stage introduce an uncertainty in position of at least 5 μm.
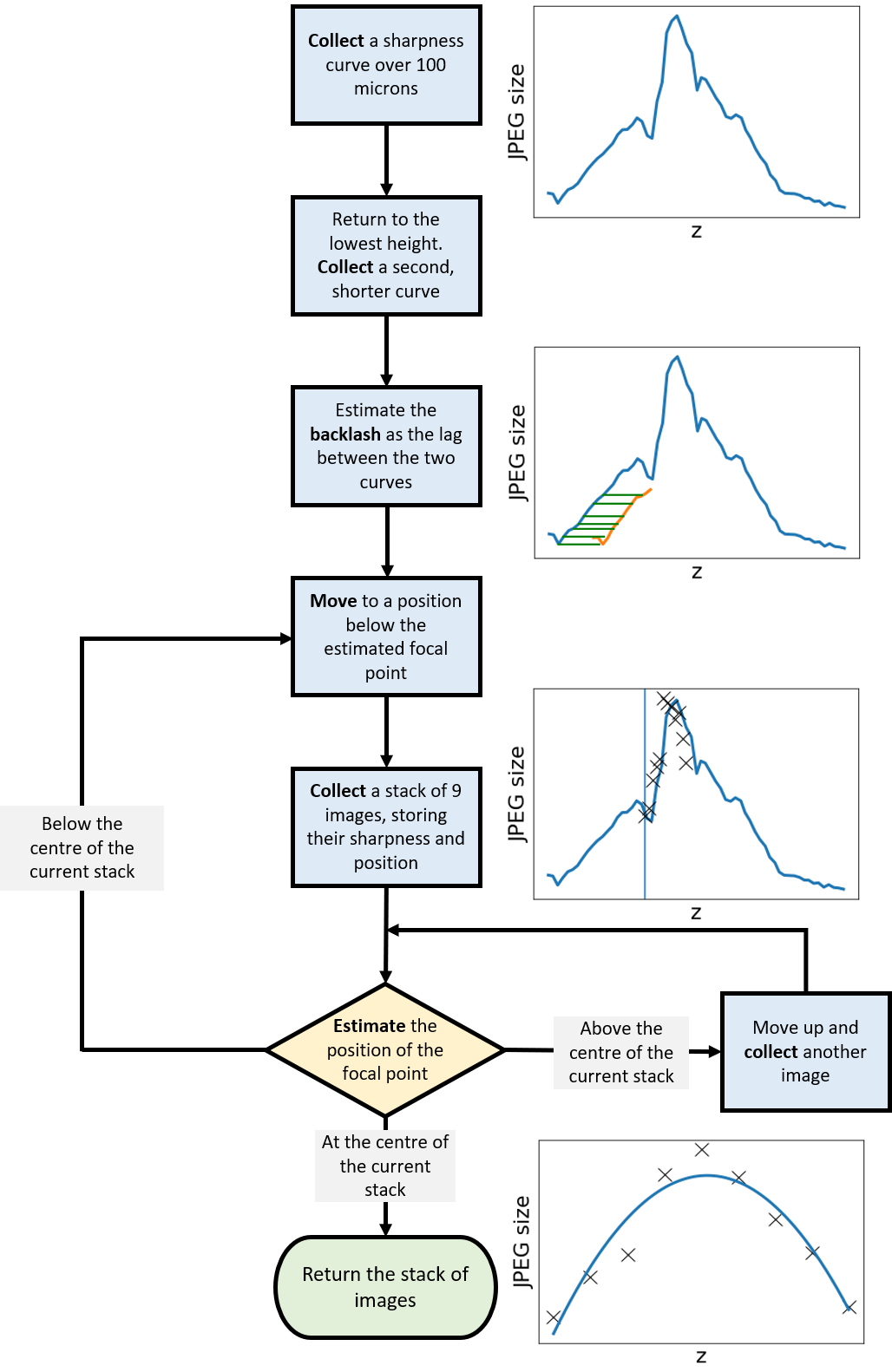
To ensure the high level of precision required for high-resolution diagnostic microscopy, we have introduced a new approach to taking z-stacks. By recording the JPEG size of images over a 100-micron range, then returning to a position far from focus, the movement required to undershoot the focal point by a suitable distance can be estimated. The microscope can then begin a ‘rolling z-stack’, capturing 9 images and assessing their size and distribution to judge the position of the stack relative to the focal point. This allows the microscope to intelligently determine success and correct its position, while ensuring the movements account for backlash and other sources of uncertainty.
Figure 3: A flowchart of the movements, measurements and captures performed in the rolling z-stack. Steps to estimate the optimal distances and intelligently abort the procedure may be added.
This change in approach is being trialled by our Tanzanian collaborators, allowing us to compare its success with our historical data. Further investigation into the optimal ranges and methods of determining success are planned. This autofocus method presents a step towards the aim of supporting low resource health clinics with automated image acquisition, by implementing a solution designed around the advantages and limitations of our setup. OpenFlexure software includes multiple options for autofocus, allowing users to select the approach most suitable for their applications.
- References
[1] Joel Collins et al. “Robotic microscopy for everyone: the OpenFlexure Microscope”
[2] Joel Collins et al. "Modern Microscopy with the Web of Things: The OpenFlexure Microscope Software Stack"
[3] Xingxing Hao, Hui Zhao, and Jing Liu. “Multifocus color image sequence fusion based on meanshift segmentation”