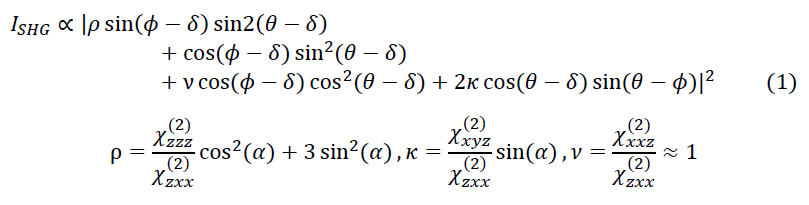Label-free imaging of collagen and myosin, using excitation polarization resolved second harmonic generation imaging through a 125 µm fiber probe
- Abstract number
- 279
- Presentation Form
- Submitted Talk
- DOI
- 10.22443/rms.mmc2021.279
- Corresponding Email
- [email protected]
- Session
- Stream 5: Label Free Imaging
- Authors
- Angel Cifuentes (2), Johanna Trägårdh (2), Tomáš Pikálek (2), Petra Ondráčková (2), Rodrigo Amezcua-Correa (1), José Enrique Antonio-Lopez (1), Tomas Čižmár (2, 3)
- Affiliations
-
1. CREOL, The College of Optics and Photonics, University of Central Florida
2. Institute of Scientific Instruments of the CAS
3. Leibniz Institute of Photonic Technology
- Keywords
Multimode fiber, SHG, microscopy, endoscope
- Abstract text
Endoscopes based on multimode fibers (MMF) have recently emerged as tools for minimally invasive imaging in tissue at depths beyond the reach of multi-photon microscopy. We here demonstrate excitation polarization-resolved second harmonic generation imaging (SHGIM) through a 125 mm diameter MMF. This allows label-free imaging and identification of non-centrosymmetric structural proteins such as collagen and myosin. We image and differentiate these proteins in mouse tail tendon and muscle.
Current clinical diagnosis of cancer and fibrotic diseases is based on performing a biopsy, followed by time-consuming histopathology off site. There is thus a need for a method for rapid in situ diagnosis, which would also reduce the need for repeated surgery. In a number of situations, in particular for tumors in hard-to-access locations such as for deep-seated brain tumors, the diagnosis method must be as minimally invasive as possible. Combining ultrathin MMF endoscopes with non-linear label-free imaging methods such as SHG and CARS that are suitable for so-called “optical biopsies”, could be a suitable solution.
Polarization resolved SHGIM [1] relies on a variable polarization state of the excitation light to probe the second-order nonlinear susceptibility tensor, c(2). c(2) depends on the sample composition, chirality and structural organization, e.g. local fibril orientation, and thus the polarization response probes these properties. In our implementation of the method a series of consecutive images, each at a different linear polarization angle are acquired. The modulation of the excitation polarization angle results in an amplitude modulation of the SHG signal, ISHG. By fitting a mathematical model to the data, and the orientation of the fibril can be probed. We used the C6 symmetry model [1] that describes the dependence of ISHG on the effective c(2); in-plane fibril orientation, 𝛿; out-of-plane fibril orientation, 𝛼; linear polarization angle of the exciting illumination, 𝜃; and the detected light polarization angle, 𝜙 :
Since GRIN fibers do not completely preserve the polarization of the transmitted light we detect and model the total intensity given by: 𝐼𝑇𝑜𝑡𝑎𝑙∝𝐼𝜙=0+𝐼 𝜙=𝜋/2
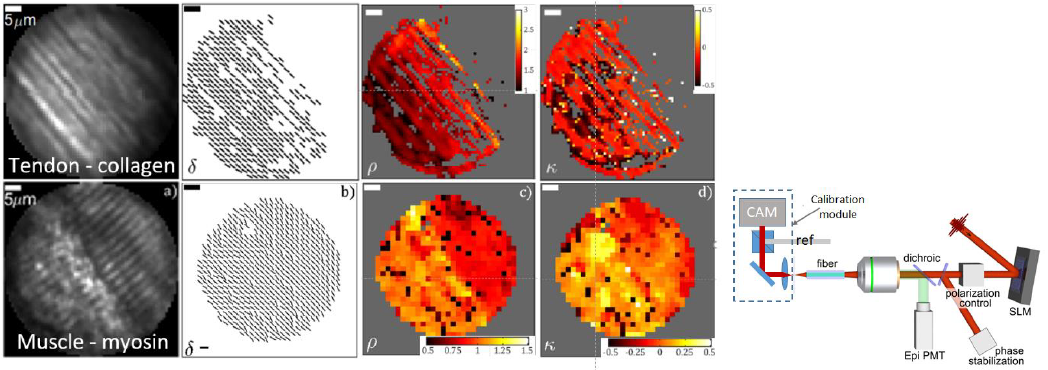
We performed polarization resolved SHGIM through a 125 mm diameter multimode GRIN fiber using a fs laser emitting at 1040 nm as excitation source (see fig 1). The details of the setup and calibration procedure has been described in [2]. In essence, the imaging is preceded by a calibration step, in which the light transport through the fiber probe is analyzed. Using this information, focal points can be created by coupling an appropriately shaped wavefront into the fiber. Each focal position corresponds to a specific pattern on the SLM and by displaying a sequence of such patterns, the focus is scanned across a plane 15 µm from the output facet of the MMF, mimicking a laser scanning microscope. In order to set the output polarization, calibrations are performed for two orthogonal linearly polarized output states (spots), by rotating the reference beam by 90° between each calibration. During imaging, the polarization state of the spot is set by setting the relative power and phase of the orthogonally polarized spots with the SLM.
The results of imaging tendon and muscle from a mouse tail are shown in figure 1. The aligned collagen fibrils in the tendon can clearly be seen both in the image and in the quiver plot for the in-plane fibril orientation d. rav=1.46, in agreement with literature values for collagen. The near 0 value for k is in line with a flat and horizontally imaged sample. For the muscle the characteristic striations are clearly visible. Here, the sample has one part consisting of pure myosin (rav =0.89) and one part where the rho value indicates an inclusion of another material, likely collagen (rav =1.06)
Figure 1. Linear polarization SHGHIM on mouse tail tendon and muscle. From left to right: sum of the intensity for all polarization angles, fibril in-plane orientation d, and parameter maps for r and k. Right: Schematic of the setup.
An issue with detecting coherently generated signals in a MMF endoscope is that the SHG signal is generated in the forward direction, and epi-detection relies on the signal being back-scattered from the tissue. The detection of such back-scattered light is limited by the size of the detector aperture, which is small for a MMF. Nevertheless, we demonstrate sufficient epi-collection efficiency for imaging.
In conclusion, we have shown that it is possible to perform SHG microscopy in a minimally invasive fashion through a MMF endoscope. We can fully control the in-plane polarization state of the scanning focus at the fiber output, and we use this to implement linearly polarized SHGIM. By imaging fixed mouse tail tendon and muscle we demonstrate that we, through the polarization modulation, achieve contrast between the structural proteins myosin and collagen at a single pixel level. Furthermore, we can detect the local orientation of the collagen fibrils. We believe that this work is a step towards a device capable of performing label-free diagnostics of tumors and fibrotic diseases in situ and at depths inaccessible to currently available microscopy techniques.
References
1. A. Golaraei et al., Journal of biophotonics 12, e201800241 (2019).
2. J. Trägårdh et al., Optics express 27, 30055-30066 (2019).
- References