The Gram-positive bacterial cell wall: an evolving heterogeneous hydrogel characterised by AFM
- Abstract number
- 357
- Corresponding Email
- [email protected]
- Session
- Stream 4 (AFM): Quantitative SPM for Biology, Biomedicine, and Bioinspired Technologies
- Authors
- Dr Laia Pasquina Lemonche (1), Dr Bartlomiej Salamaga (1), Prof Simon Foster (1), Prof Jamie Hobbs (1)
- Affiliations
-
1. University of Sheffield
- Keywords
AFM, High-resolution, Bacterial cells, Peptidoglycan, Antibiotics, Image Analysis, Biophysics
- Abstract text
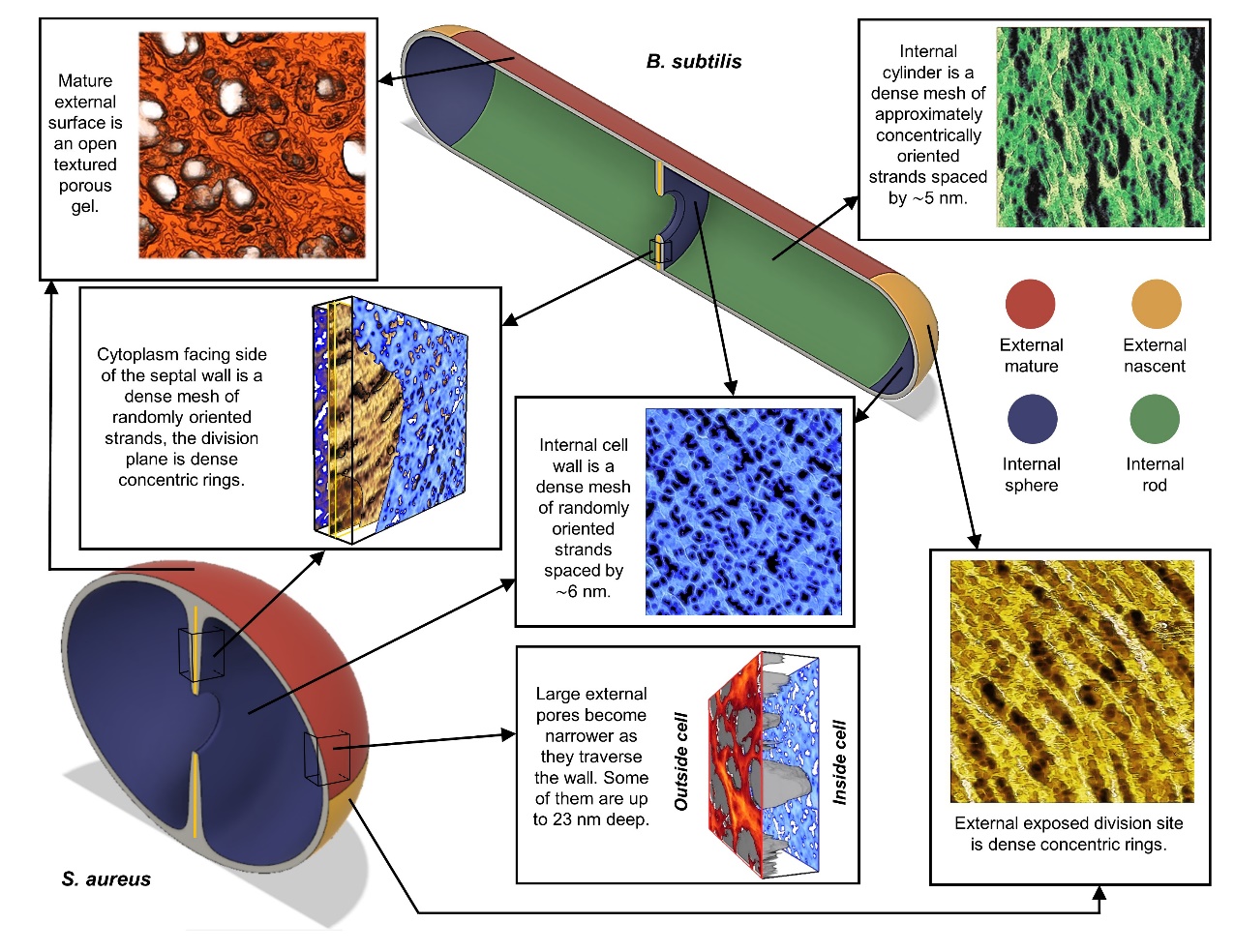
The primary structural component of the bacterial cell wall is peptidoglycan which is crucial for survival and division of the cells. Peptidoglycan (PG) is a heterogeneous macromolecule composed of glycan chains (sugars) and small peptides that unify the structure by crosslinking the glycan chains. PG is tens of nanometres thick in Gram-positive bacteria and acts as a constraint to interrupt turgor. This component of the cell is also one of the major targets by cell wall antibiotics such as Methicillin or Vancomycin.
In this project, we applied several imaging modes of atomic force microscopy (AFM) to interrogate the morphology of this heterogeneous hydrogel. The bacteria of study were Staphylococcus aureus and Bacillus subtilis which are two Gram-positive species with distinct cell shape (sphere vs rod). High resolution tapping was used to image the external surface of live cells and PeakForce Tapping was used to study both the internal and external surface of purified PG. Then, quantitative image analysis methods were developed to obtain robust conclusions when comparing different samples. The results was that contrary to stablished theories, the PG is not an homogeneous impenetrable wall, it is a highly porous heterogeneous hydrogel [1].
Then, once the architecture of PG from healthy cells was well characterised, the same techniques were applied to study the effect of different antibiotics (Methicillin and Vancomycin) to the PG morphology. We used Staphylococcus aureus wild type, together with different mutants to decipher the role of PG modification enzymes during cell death [2]. The results corroborate the model that for the cell to survive there must be an equilibrium between synthesis and hydrolisis of PG, this equilibrium is disrupted when antibiotics are applied, therefore leading to cell death. This brings us one step closer to obtain a complete bigger picture of how the bacterial cell wall evolves during the life and death cycle.
- References
[1] L. Pasquina-Lemonche, J. Burns, R.D. Turner, S. Kumar, R. Tank, N. Mullin, J. S. Wilson, B. Chakrabarti, P. A. Bullough, S. J. Foster, J. K. Hobbs, “The architecture of the Gram-positive bacterial cell wall”, Nature, 582, 294-297 (2020).
[2] Bartlomiej Salamaga, Lingyuan Kong, Laia Pasquina-Lemonche, Lucia Lafage, Milena von und zur Muhlen, Josie F. Gibson, Danyil Grybchuk, Amy Tooke, Viralkumar Panchal, Elizabeth J. Culp, Elizabeth Tatham, Mary E. O’Kane, Thomas E. Catley, Stephen A. Renshaw, Gerard D. Wright, Pavel Plevka, Per A. Bullough, Aidong Han, Jamie K. Hobbs, Simon J. Foster, “Demonstration of the role of cell wall homeostasis in Staphylococcus aureus growth and the action of bactericidal antibiotics”, PNAS, submitted (2021)